Advanced Example cellularPhantom
Responsible Geant4 Collaborator: Sebastien Incerti, CNRS, Bordeaux U., LP2I Bordeaux, France.
Developers: P. Barberet (a), S. Incerti (a), N. H. Tran (a), L. Morelli (a,b):
- a) CNRS, Bordeaux U., LP2I Bordeaux, France
- b) Politecnico di Milano, Italy
Introduction
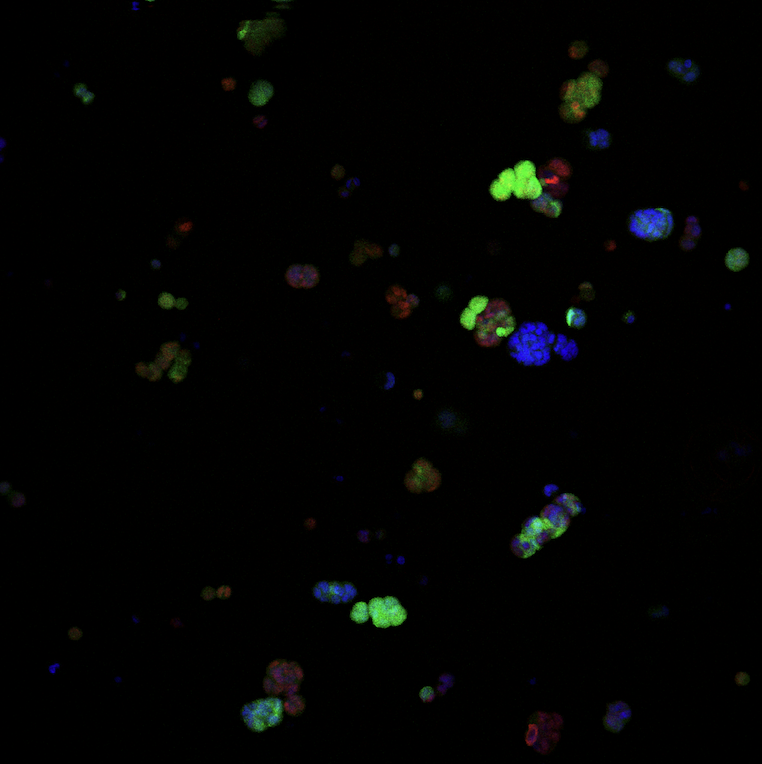
The cellularPhantom example shows how to simulate the irradiation of a 3D voxel phantom containing several biological cells, created from a confocal microscopy 24-bit RGB image.
The example provides two phantoms (low and high resolution), both obtained from an image (see Figure 1) that was created thanks to:
- H. De Oliveira, T. Désigaux, N. Dusserre - ART BioPrint, France
- F. Paris, C. Niaudet - Inserm, France
These developments were carried out as part of the “Flash’Atlantic” project (2023-2024) funded by CNRS-MITI, France, and Inserm, France.
Phantoms
The two phantom files, phantom.dat (low resolution) and phantomHR.dat (high resolution), are provided (automatic download upon compilation and located in the phantoms directory). They were created using the ImageJ phantom.ijm macro, which is located in the ImageJ directory. See the Documentation.pdf file (automatic download upon compilation and located in the phantoms directory) for more information on how to create such phantom files.
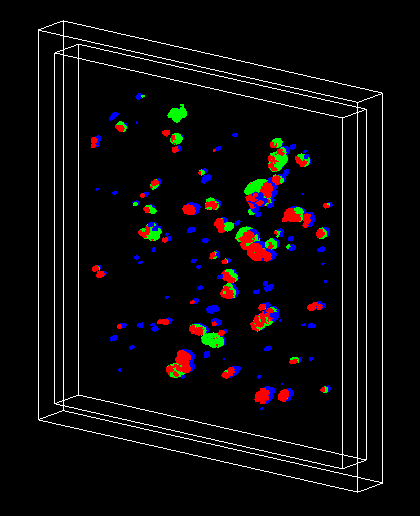
The low-resolution file is used for visualization in the macro vis.mac (see Figure 2). It contains the following lines:
54300 20230 17320 16750 => total number of voxels, number of red, green and blue voxels
734.0507 734.0507 90.6372 microns => whole X, Y and Z size of the phantom, with unit
2.8674 2.8674 2.0142 microns => size of a single voxel, with unit
and the list of individual voxels, with the format: X, Y and Z positions, type (type is 1 for red, 2 for green, 3 for blue): 232.2582 31.5412 0.0000 2 235.1256 31.5412 0.0000 2 …
The low-resolution and high-resolution files can be used by the run.mac macro.
How to run the simulation
The geometry is a 1-mm side cube (“World”) made of air, with a thickness of 100 um, containing a liquid water medium (“Medium”) of side 900 um and thickness 95 um, containing itself the phantom (“Phantom”). The World and Medium dimensions can be changed by UI command.
The PhysicsList class uses Geant4 option4 electromagnetic physics. It also contains other physics lists including Geant4-DNA option2, which is commented by default.
The simulation is controlled by the run.mac macro file. In this macro, the user can select:
- the number of threads (MT mode)
- the phantom file name
- the World and Medium dimensions
- the Medium material
- the phantom voxel density
- the position (shift in X or Y or Z) of the phantom in the Medium
- the production cuts outside and inside in the phantom
- the incident particles (using GPS)
Simulation results
The following results are displayed at the end of the simulation:
- total number of voxels in phantom
- total number of RED voxels in phantom
- total number of GREEN voxels in phantom
- total number of BLUE voxels in phantom
- total absorbed energy in RED voxels (MeV)
- total absorbed energy in GREEN voxels (MeV)
- total absorbed energy in BLUE voxels (MeV)
- total absorbed dose in RED voxels (Gy)
- total absorbed dose in GREEN voxels (Gy)
- total absorbed dose in BLUE voxels (Gy)
A phantom.root result file contain three ntuples, corresponding to the 3 types of voxels (red, green and blue) of the original image. The following voxel information is available in these ntuples:
- x, y, z position
- energy deposition
- absorbed dose
- voxel number (ID)
The ROOT macro plot.C can be run to display:
- the cellular phantom
- the absorbed energy distribution in the 3 types of voxels
- the absorbed energy 2D map for the 3 types of voxels
- the absorbed dose 2D map for the 3 types of voxels
Simply do, after the simulation: root plot.C
References
Monte-Carlo dosimetry on a realistic cell monolayer geometry exposed to alpha-particle, P. Barberet, F. Vianna, M. Karamitros, T. Brun, N. Gordillo, P. Moretto, S. Incerti, H. Seznec, Phys. Med. Biol. 57 (2012) 2189-2207 link
Monte Carlo dosimetry for targeted irradiation of individual cells using a microbeam facility, S. Incerti, H. Seznec, M. Simon, Ph. Barberet, C. Habchi, Ph. Moretto, Rad. Prot. Dos. 133, 1 (2009) 2-11 link
Last updated: 23/12/2024 by S. Incerti